-
Notifications
You must be signed in to change notification settings - Fork 0
Transcript Assembly Visualization
#4-v. Transcript Assembly Visualization (Splicing Visualization)
View the junctions.bed files created by TopHat
cd $RNA_HOME/alignments/tophat/UHR_Rep1_ERCC-Mix1/
head junctions.bed
View the merged GTF file from the 'de_novo' mode. Remember this merged GTF file combines both UHR and HBR (GTFs for each individually were also produced earlier).
cd $RNA_HOME/expression/tophat_cufflinks/de_novo/merged/
head merged.gtf
For details on the format of these files, refer to the following links:
- http://cole-trapnell-lab.github.io/cufflinks/cuffmerge/index.html
- http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/index.html#transfrag-class-codes
View the differential splicing, differential promoter usage and differential CDS results files. For each results file, we sort the results by the p-value and view the top 10 isoforms, ignoring entries classified as 'LOWDATA'.
cd $RNA_HOME/de/tophat_cufflinks/de_novo/
sort -k 12n splicing.diff | grep -v LOWDATA | head
sort -k 12n promoters.diff | grep -v LOWDATA | head
sort -k 12n cds.diff | grep -v LOWDATA | head
How many genes have at least one transcript assembled by Cufflinks in the 'de_novo' results?
cd $RNA_HOME/expression/tophat_cufflinks/de_novo/merged/
cat merged.gtf | perl -ne 'if ($_ =~ /gene_name\s\"(\w+)\"/){print "$1\n"}' | sort | uniq | wc -l
How many genes have at least one novel transcript assembled?
grep "j" merged.gtf | perl -ne 'if ($_ =~ /gene_name\s\"(\w+)\"/){print "$1\n"}' | sort | uniq | wc -l
##Visualizing Results in the IGV Browser
###junctions.bed files:
-
View the tophat junctions.bed file (generated all the way back in Module 3)
-
The following can be loaded directly in IGV by url
-
http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/UHR_Rep1_ERCC-Mix1/junctions.bed
-
http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/HBR_Rep1_ERCC-Mix2/junctions.bed
-
Go to the gene 'RAC2'
-
Do you see the evidence for any novel exons?
###merged.gtf files:
- View the grand merged.gtf files that were generated by each of the three Cufflinks modes: 'ref_only', 'ref_guided', 'de_novo'.
- The following can be loaded directly in IGV by url
- http://YOUR_IP_ADDRESS/workspace/rnaseq/expression/tophat_cufflinks/ref_only/merged/merged.gtf
- http://YOUR_IP_ADDRESS/workspace/rnaseq/expression/tophat_cufflinks/ref_guided/merged/merged.gtf
- http://YOUR_IP_ADDRESS/workspace/rnaseq/expression/tophat_cufflinks/de_novo/merged/merged.gtf
Load the BAM files at the same time as the junctions.bed and merged.gtf files:
- The following can be loaded directly in IGV by url
- http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/UHR_ERCC-Mix1_ALL/accepted_hits.bam
- http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/HBR_ERCC-Mix2_ALL/accepted_hits.bam
Go to the following regions:
- 22:45730787-45736825
- 22:45607463-45610990
Do you see the evidence for any novel exons/transcript that are found in 'de_novo' or 'ref_guided' modes but NOT found in 'ref_only' mode? Explore in IGV for other examples of novel or different transcript predictions from the different cufflinks modes. Pay attention to how the predicted transcripts line up with known transcripts. Try loading the Ensembl transcripts track (File -> Load from Server).
NOTE: We have obviously just scratched the surface exploring these output files.
##SAVING A COPY OF YOUR RESULTS TO TAKE HOME WITH YOU If you are performing this tutorial on a cloud instance, everything will be deleted when the instance is destroyed! To package and download everything used or created during the tutorials you can do the following from your cloud terminal session.
First package and compress all of the directories and files in the ‘rnaseq’ directory
cd /home/ubuntu/workspace/
tar -czvf rnaseq_tutorial.tar.gz rnaseq/
Now you can download this to your own computer from here:
- http://YOUR_IP_ADDRESS/workspace/rnaseq_tutorial.tar.gz
To unpack this archive at a terminal session on your own Linux or Mac computer you can do the following:
tar -xzvf rnaseq_tutorial.tar.gz
| Previous Section | This Section | Next Section | |:-----------------------------------------------:|:------------------------------------------------------------:|:-------------------------:| | Differential Splicing | Splicing Visualization | Kallisto |
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
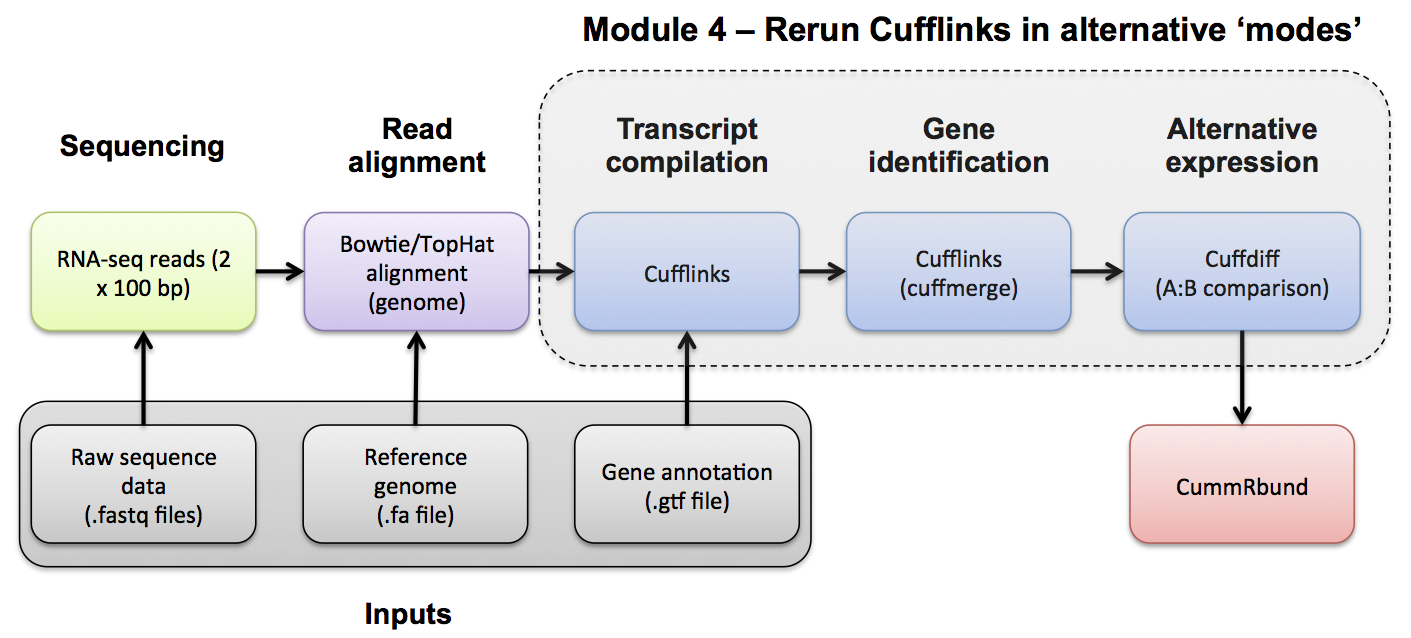
Module 4: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 5: Kallisto
Appendix: Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup