-
Notifications
You must be signed in to change notification settings - Fork 0
PostAlignment QC
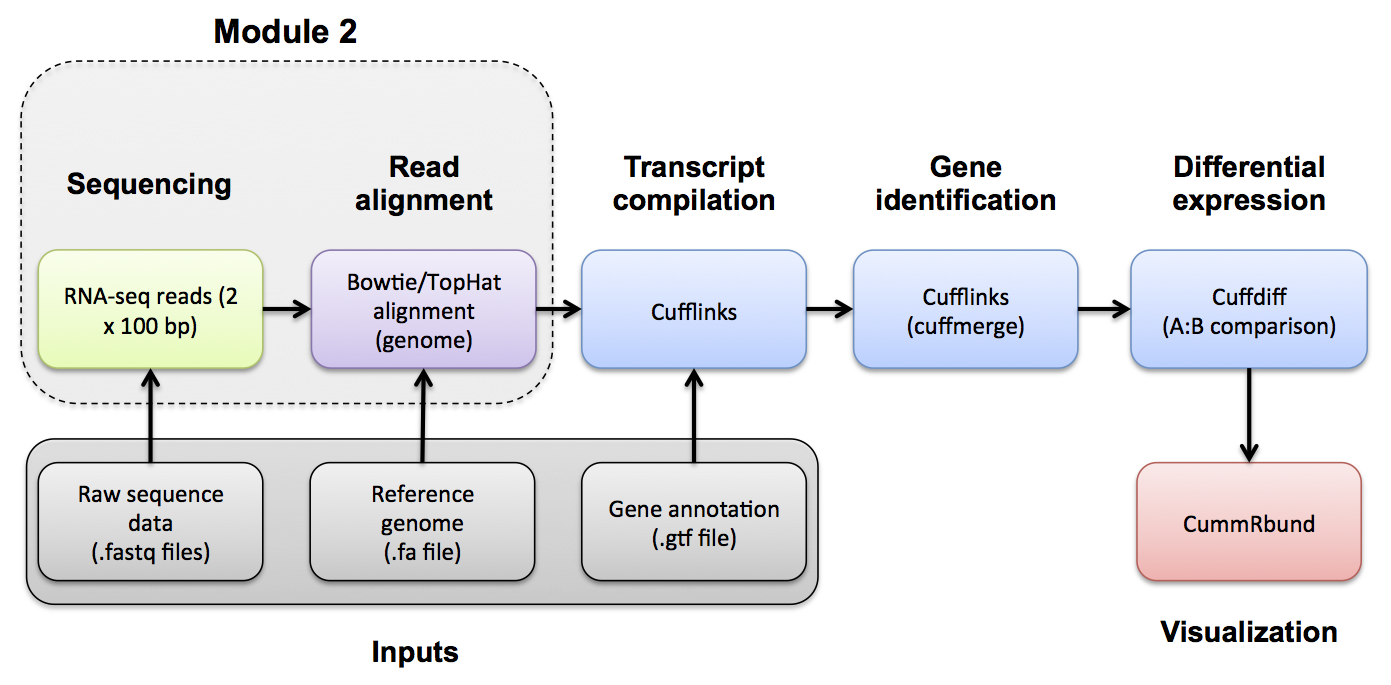
#2-iv. Post-Alignment QC ##Use samtools, samstat, and FastQC to evaluate the alignments
Use samtools view to see the format of a SAM/BAM alignment file
cd $RNA_HOME/alignments/tophat/UHR_ERCC-Mix1_ALL
samtools view -H accepted_hits.bam
samtools view accepted_hits.bam | head
Try filtering the BAM file to require or exclude certain flags. This can be done with samtools view -f -F options
-f INT required flag -F INT filtering flag
"Samtools flags explained"
Try requiring that alignments are 'paired' and 'mapped in a proper pair' (=3). Also filter out alignments that are 'unmapped', the 'mate is unmapped', and 'not primary alignment' (=268)
samtools view -f 3 -F 268 accepted_hits.bam | head
Now require that the alignments be only for 'PCR or optical duplicate'. How many reads meet this criteria? Why?
samtools view -f 1024 accepted_hits.bam | head
Use samtools flagstat to get a basic summary of an alignment. What percent of reads are mapped? Is this realistic? Why?
cd $RNA_HOME/alignments/tophat/
samtools flagstat UHR_ERCC-Mix1_ALL/accepted_hits.bam
samtools flagstat HBR_ERCC-Mix2_ALL/accepted_hits.bam
Run samstat on UHR/HBR BAMs
mkdir ~/bin
cd ~/bin
wget http://genome.wustl.edu/pub/rnaseq/tools/bin/samstat
chmod +x samstat
cd $RNA_HOME/alignments/tophat/
~/bin/samstat UHR_ERCC-Mix1_ALL/accepted_hits.bam
~/bin/samstat HBR_ERCC-Mix2_ALL/accepted_hits.bam
View the samstat summary file in a web browser. Note, you must replace YOUR_IP_ADDRESS with your own amazon instance IP address:
- http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/UHR_ERCC-Mix1_ALL/accepted_hits.bam.html
- http://YOUR_IP_ADDRESS/workspace/rnaseq/alignments/tophat/HBR_ERCC-Mix2_ALL/accepted_hits.bam.html
Details of the SAM/BAM format can be found here: http://samtools.sourceforge.net/SAM1.pdf
##Using FastQC You can use FastQC to perform basic QC of your accepted_hits.bam file (See Pre-Alignment QC). This will give you output very similar to when you ran FastQC on your fastq files.
Background: RSeQC is a tool that can be used to generate QC reports for RNA-seq. For more information, please check: RSeQC Tool Homepage
Objectives: In this section, we will try to generate a QC report for a data set downloaded from RSeQC website.
Files needed:
- Aligned bam file.
- Index file for the aligned bam.
- A RefSeq bed file.
Copy RSeQC Data
Set your working directory and copy the necessary files
cp -r ~/CourseData/RNA_data/RSeQC/RSeQC.zip ~/workspace/rnaseq/tools/
cd ~/workspace/rnaseq/tools/
Unzip the RSeQC file:
unzip RSeQC.zip
cd RSeQC/
Note: You should now see the bam, index, and RefSeq bed files listed. The bam file here is an pair-end non-strand specific example dataset from the RSeQC website.
Run RSeQC commands:
bam_stat.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam
clipping_profile.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial -s "PE"
geneBody_coverage.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
infer_experiment.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam
inner_distance.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
junction_annotation.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
junction_saturation.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
read_distribution.py -r hg19_RefSeq.bed -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam
read_duplication.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
read_GC.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
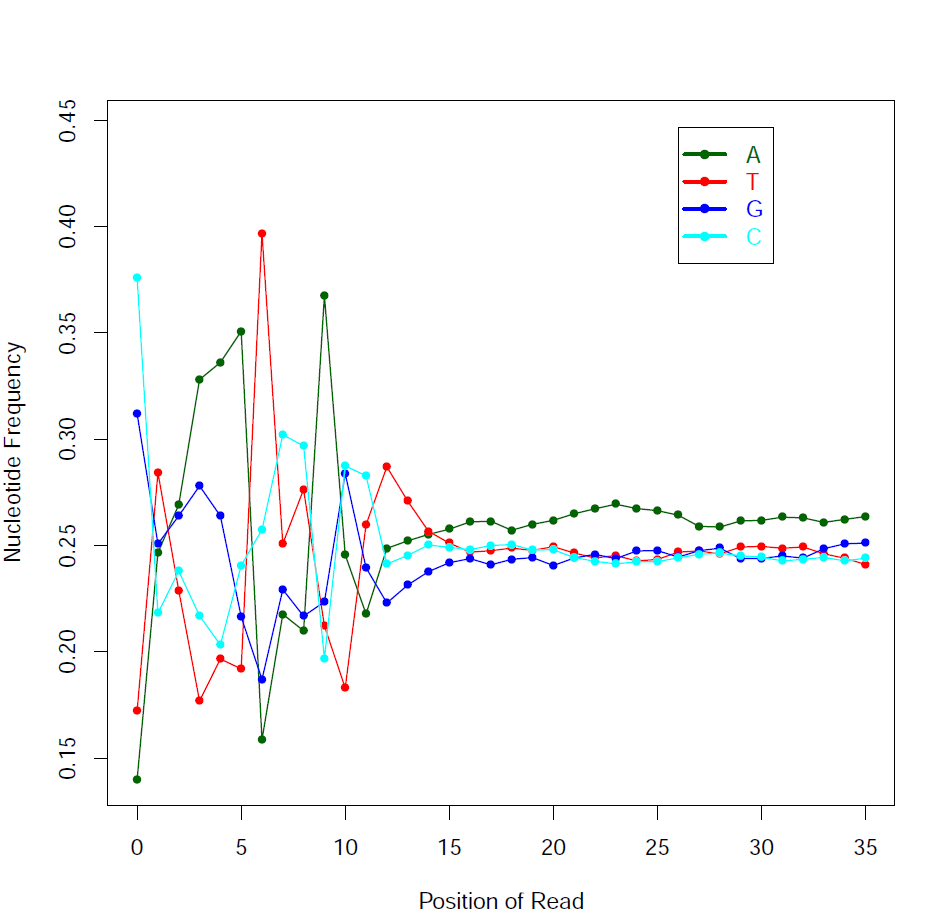
read_NVC.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
read_quality.py -i Pairend_nonStrandSpecific_36mer_Human_hg19.bam -o tutorial
ls *.pdf
Go through the generated PDFs by browsing through the following directory in a web browser:
- http://YOUR_IP_ADDRESS/workspace/rnaseq/tools/RSeQC/
Read Quality:
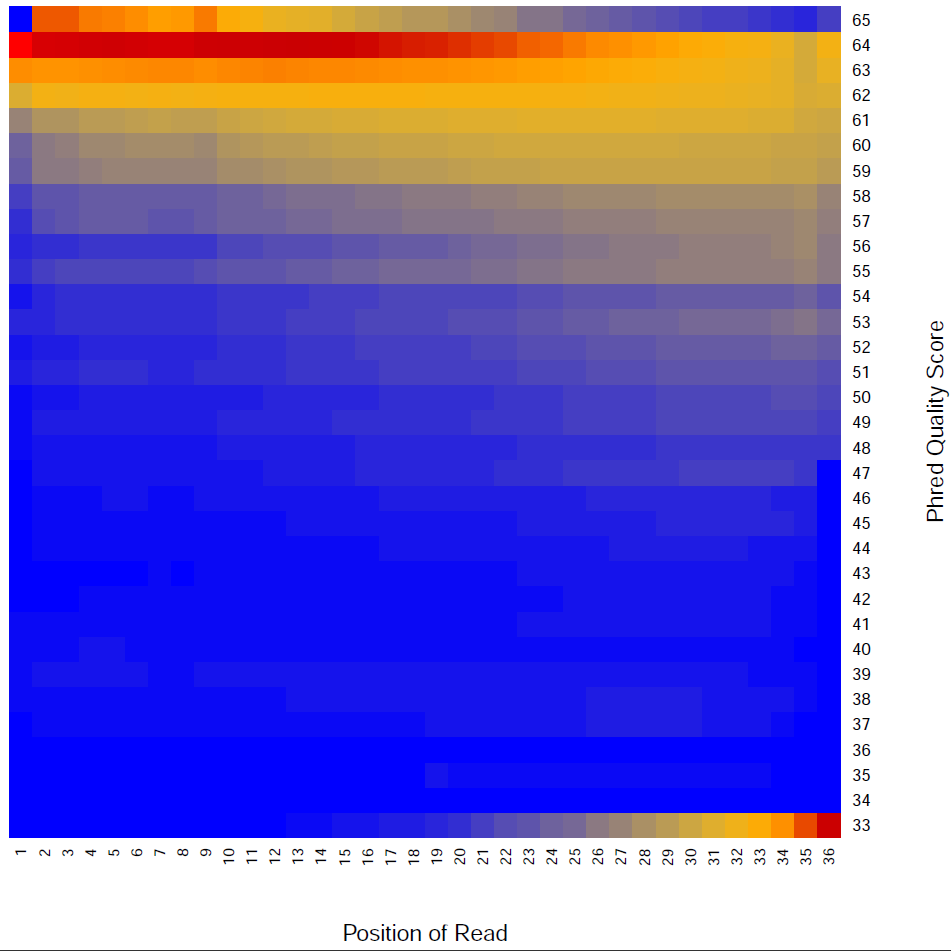
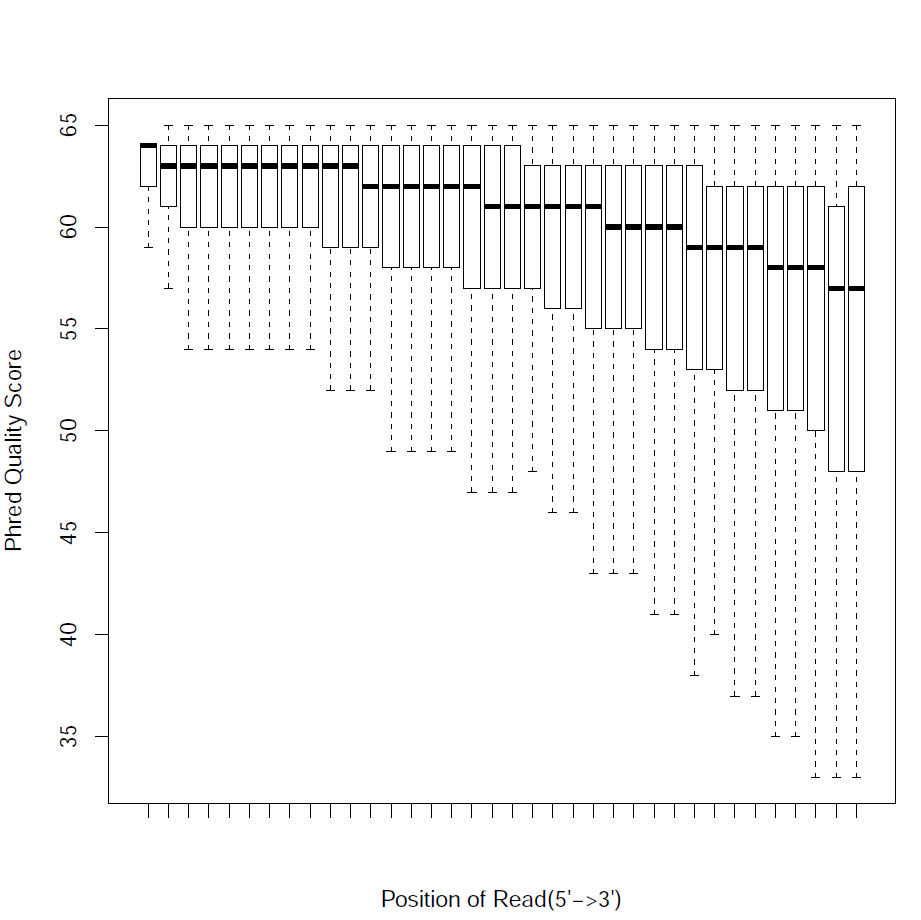
Read - Nucleotide vs Cycle (Phred base score vs. position in read):
The pattern we see here at the beginning of the reads may be caused by biases caused by random hexamer priming that arose when making cDNA from RNA (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2896536/ for further discussion).
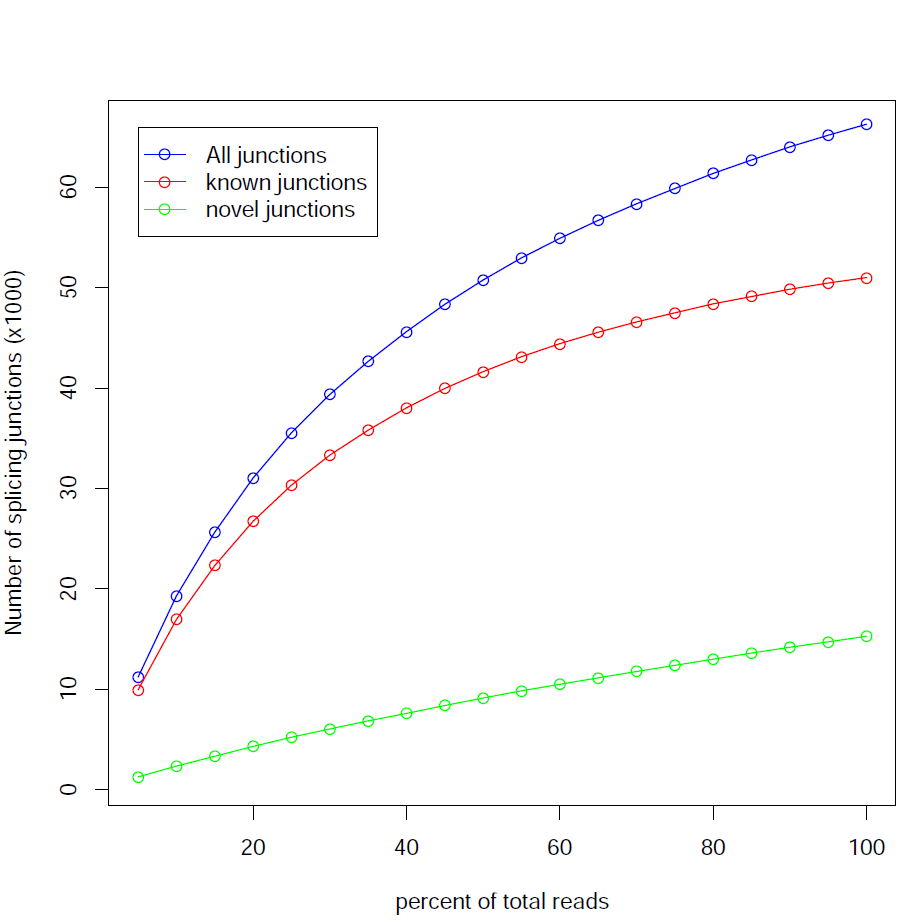
Junction Saturation:
This module checks for saturation of junction discovery by resampling 5%, 10%, 15%, ..., 95% of total alignments from the BAM or SAM file. The number of junctions discovered at each level of downsampling is plotted. If we are exhausting the junction information in our library, the line will plateau as the amount of data increases.
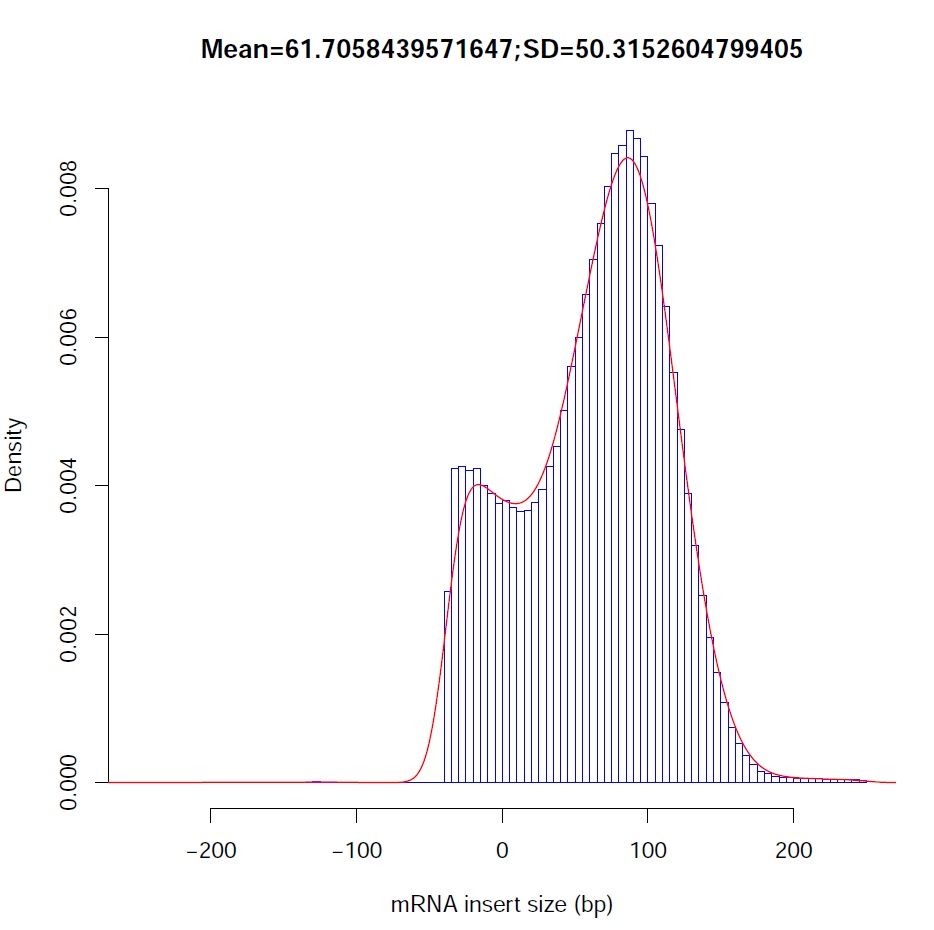
Distribution of observed inner distances in fragments:
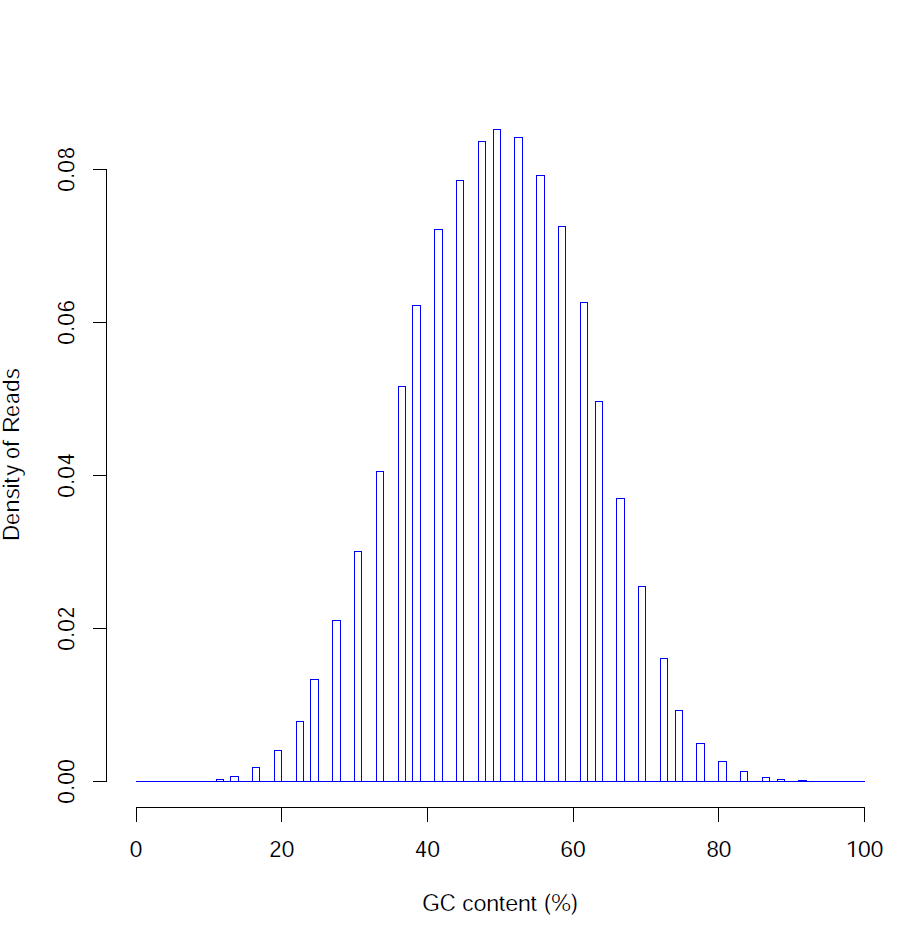
Distribution of read GC content (%):
| Previous Section | This Section | Next Section | |:-------------------------------------------------------:|:---------------------------------:|:---------------------------------------------:| | Alignment Visualization | Alignment QC | Expression |
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
Module 4: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 5: Kallisto
Appendix: Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup