Repository that contain the main data and scripts to recompute the figures from:
De Anda V, Zapata Peñasco I, Blaz J, Poot-Hernandez AC, Contreras Moreira B, Gonzales Laffite Marcos, Hernandez Rosales M, Gamez Tamariz N, Eguiarte Fruns E, Souza V. Understanding the mechanisms behind the response to environmental perturbations in microbial mats: a metagenomic-network based approach Front. Microbiol. 9:2606. doi: 10.3389/fmicb.2018.02606
Last updated: 11/29/2018
To reproduce the figures first install jupyter notebook and then clone the repository.
git clone https://github.com/valdeanda/Time_series_mats.git
cd scripts
ipython3 notebook In order to reproduce all the figures the following libraries and packages must be installed first
Python
- Pandas >= 0.20.3
- Numpy >= 1.13.3
- Seaborn >= 0.8.0
- Matplotlib >= 2.02
R packages
- Phyloseq
- ggplot2
- vegan
- pvclust
- devtools
- fpc
-
To compute the ecological indexes plase open the following script directly in your browser.
-
To obtain the final figure
The script is available in notebook format
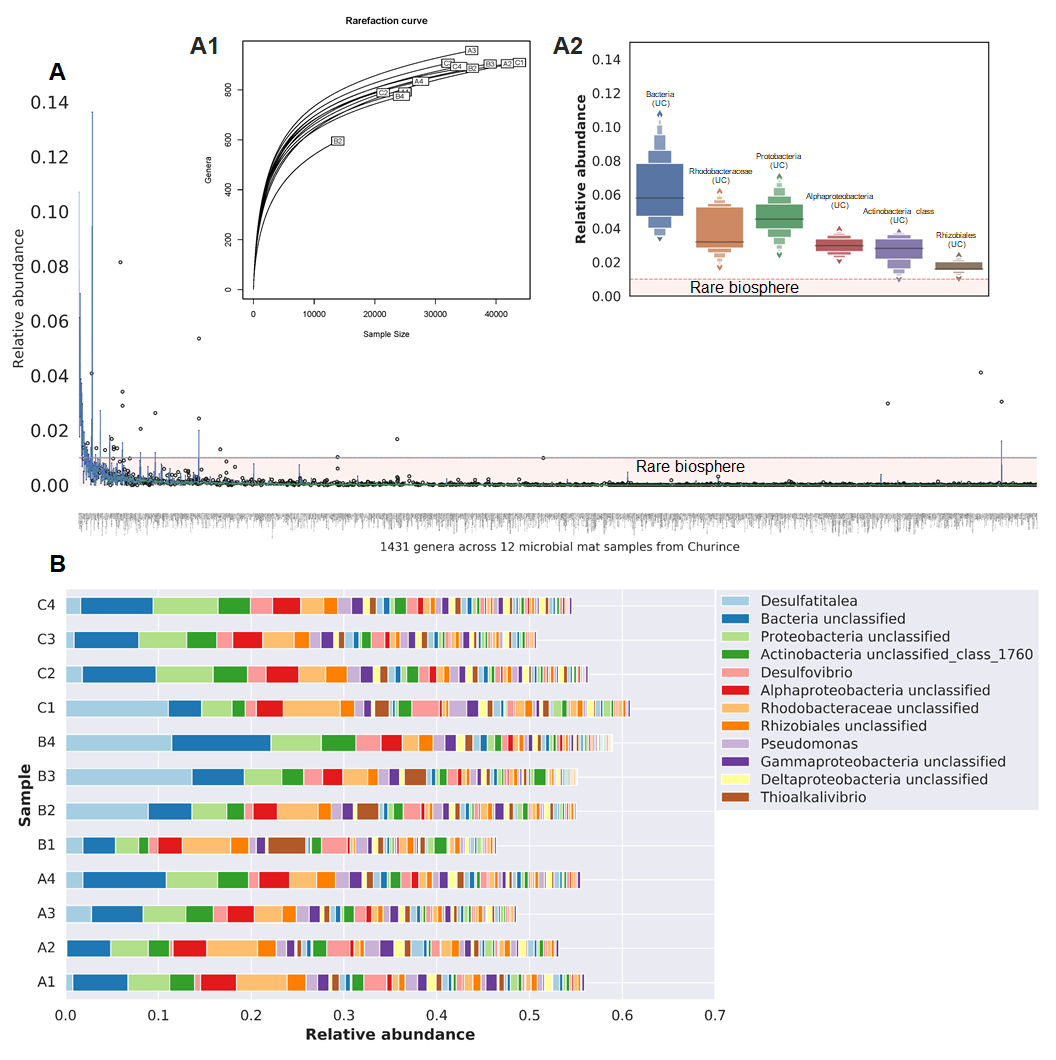
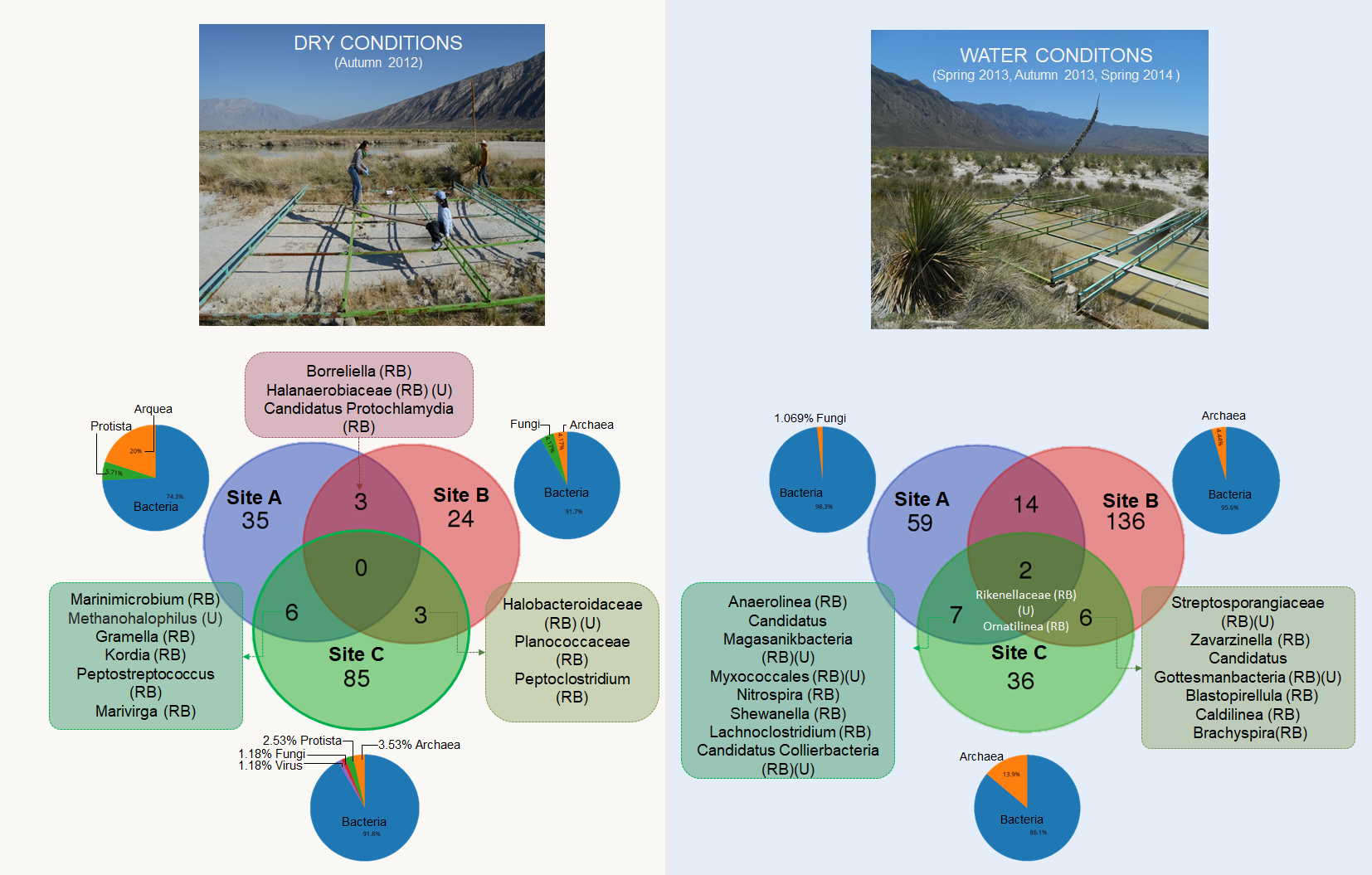
jupyter notebook Diversity.ipynb- To compute the microbial mat core and the presence absence profile of each site we used get_homologues. The script is available in notebook format. To repeat the analysis please type in your terminal:
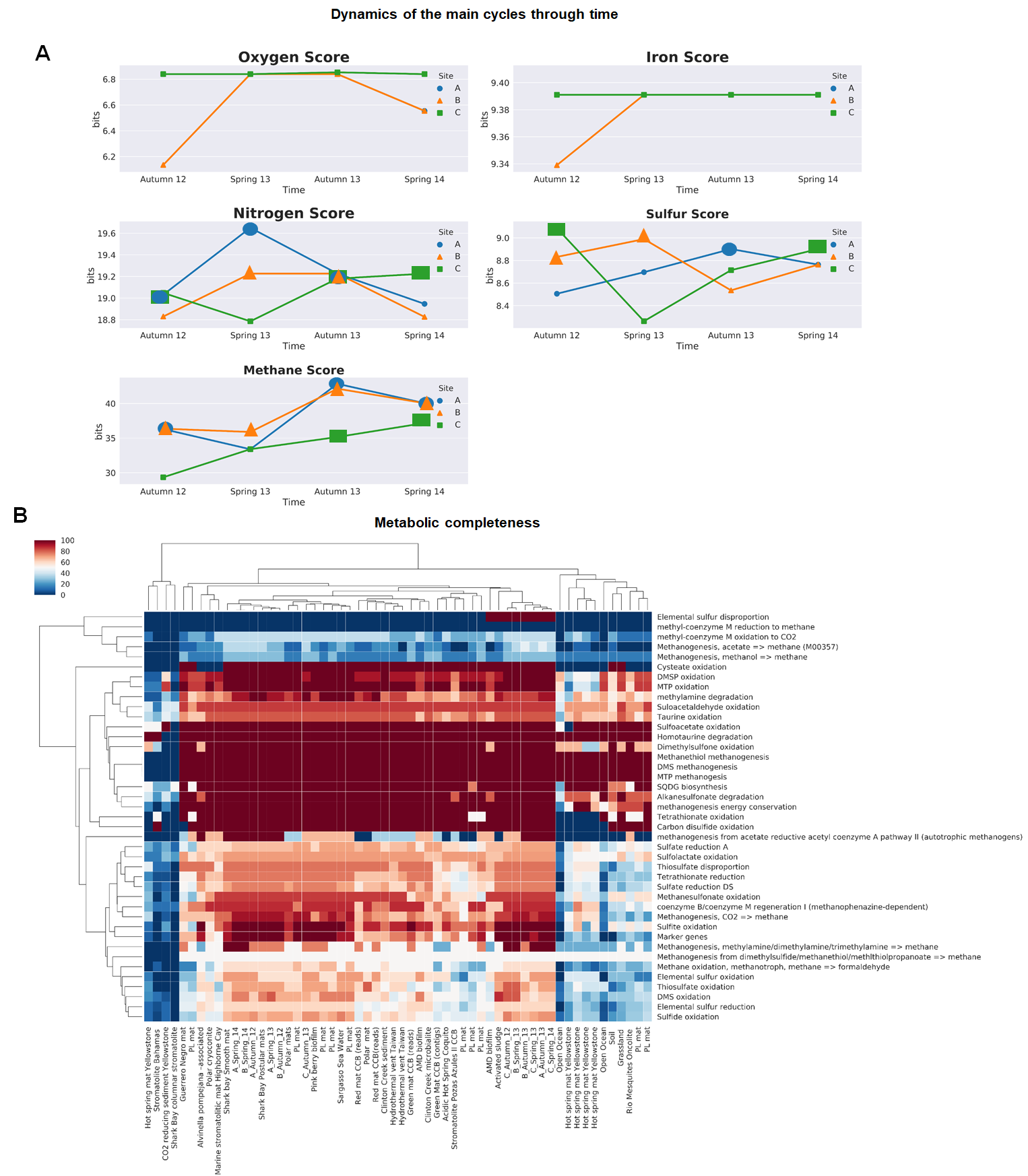
jupyter notebook Core_bacteria.ipynb- Frist compute MEBS with the following script in notebook format.
jupyer notebook MebsInTime.ipynb- Then compute the metbolic completeness
Script available in notebook format.
jupyter notebook completeness.ipynbOpen the following script directly in R
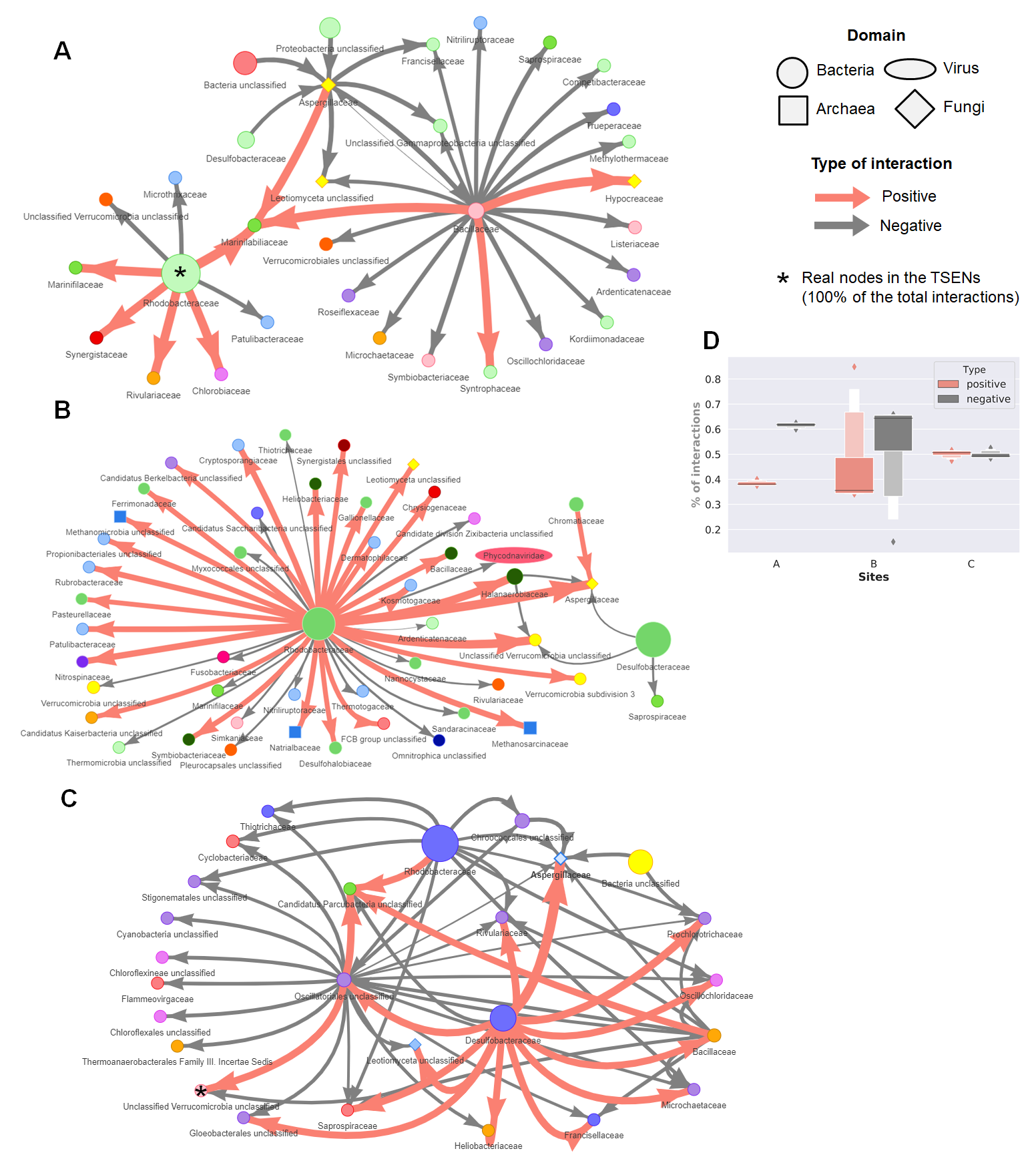
The dynamic networks for each site are available here:
Script available in notebook format.
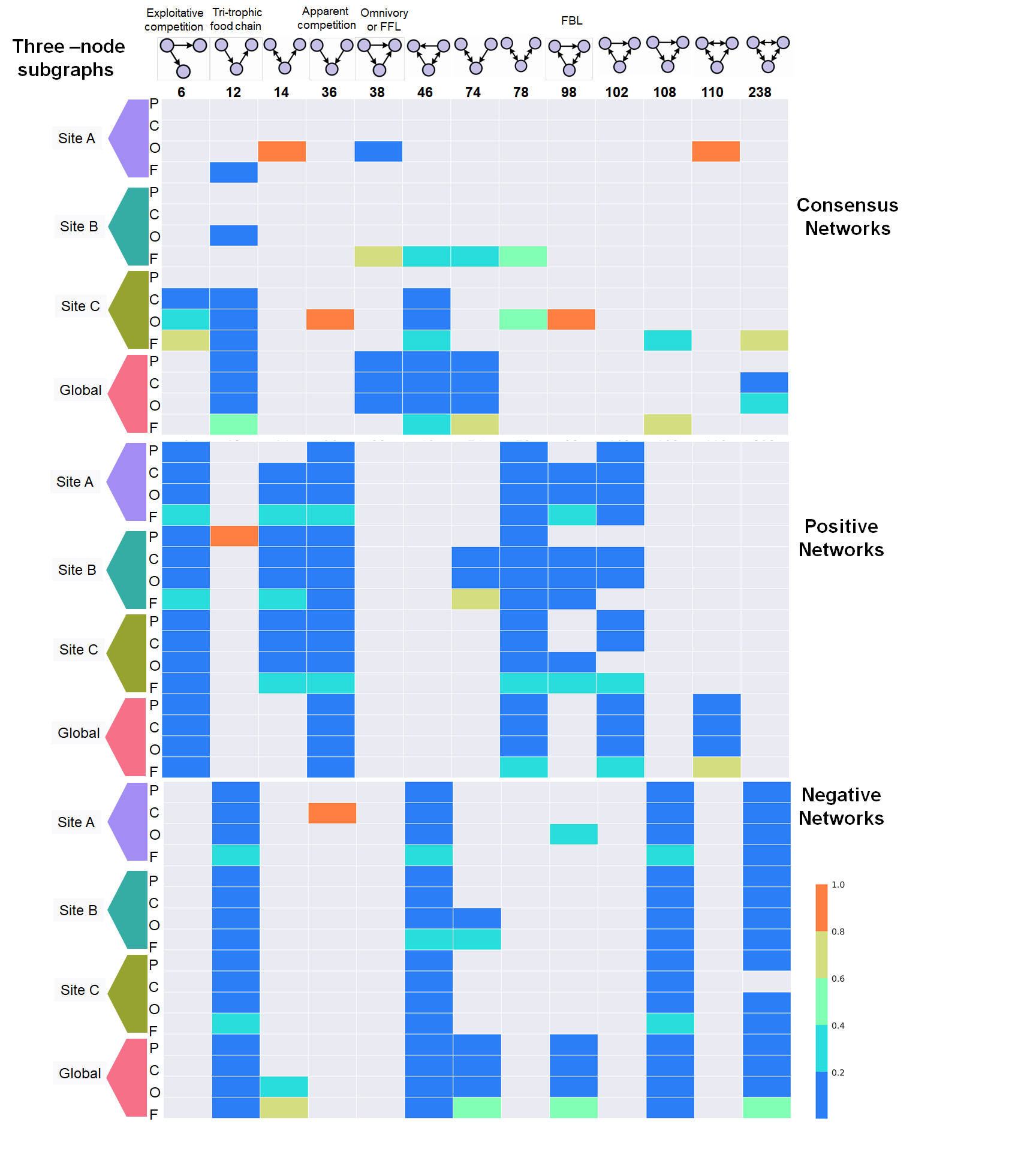
jupyter notebook Motifs.ipynbFor more information about Cuatro Cienegas please visit the documentary film, click the image below