The proffer package profiles R code to find bottlenecks. Visit
https://r-prof.github.io/proffer/ for documentation.
https://r-prof.github.io/proffer/reference/index.html has a complete
list of available functions in the package.
This data processing code is slow.
system.time({
n <- 1e5
x <- data.frame(x = rnorm(n), y = rnorm(n))
for (i in seq_len(n)) {
x[i, ] <- x[i, ] + 1
}
x
})
#> user system elapsed
#> 82.060 28.440 110.582 Why exactly does it take so long? Is it because for loops are slow as
a general rule? Let us find out empirically.
library(proffer)
px <- pprof({
n <- 1e5
x <- data.frame(x = rnorm(n), y = rnorm(n))
for (i in seq_len(n)) {
x[i, ] <- x[i, ] + 1
}
x
})
#> ● url: http://localhost:57517
#> ● host: localhost
#> ● port: 57517When we navigate to http://localhost:64610 and look at the flame
graph, we see [<-.data.frame() (i.e. x[i, ] <- x[i, ] + 1) is taking
most of the runtime.
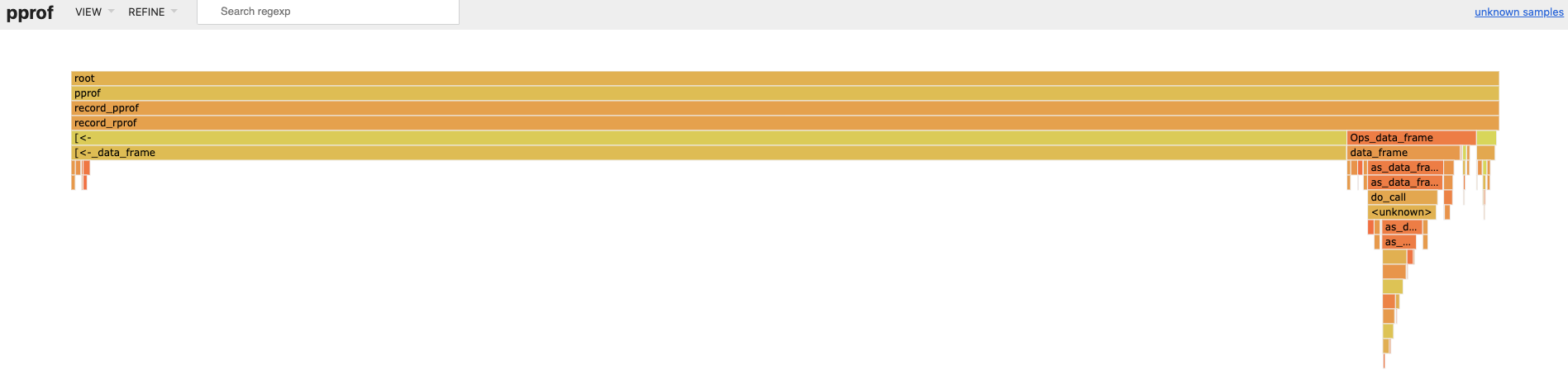
So we refactor the code to avoid data frame row assignment. Much faster,
even with a for loop!
system.time({
n <- 1e5
x <- rnorm(n)
y <- rnorm(n)
for (i in seq_len(n)) {
x[i] <- x[i] + 1
y[i] <- y[i] + 1
}
x <- data.frame(x = x, y = y)
})
#> user system elapsed
#> 0.019 0.001 0.020Moral of the story: before you optimize, throw away your assumptions and run your code through a profiler. That way, you can spend your time optimizing where it counts!
The pprof server is a background
processx process, and you can
manage it with the processx methods described
here. Remember
to terminate the process with $kill() when you are done with it.
# px is a process handler.
px <- pprof({
n <- 1e4
x <- data.frame(x = rnorm(n), y = rnorm(n))
for (i in seq_len(n)) {
x[i, ] <- x[i, ] + 1
}
x
})
#> ● url: http://localhost:50195
#> ● host: localhost
#> ● port: 50195
# Summary of the background process.
px
#> PROCESS 'pprof', running, pid 10451.
px$is_alive()
# [1] TRUE
# Error messages, some of which do not matter.
px$read_error()
#> [1] "Main binary filename not available.\n"
# Terminate the process when you are done.
px$kill()As with Jupyter notebooks, you can serve pprof from one computer and
use it from another computer on the same network. On the server, you
must
- Find the server’s host name or IP address in advance.
- Supply
"0.0.0.0"as thehostargument.
system2("hostname")
#> mycomputer
px <- pprof({
n <- 1e4
x <- data.frame(x = rnorm(n), y = rnorm(n))
for (i in seq_len(n)) {
x[i, ] <- x[i, ] + 1
}
x
}, host = "0.0.0.0")
#> ● url: http://localhost:610712
#> ● host: localhost
#> ● port: 610712Then, in the client machine navigate a web browser to the server’s host
name or IP address and use the port number printed above,
e.g. https://mycomputer:61072.
For old versions of proffer (0.0.2 and below) refer to these older
installation
instructions
instead of the ones below.
The latest release of proffer is available on
CRAN.
install.packages("proffer")Alternatively, you can install the development version from GitHub.
# install.packages("remotes")
remotes::install_github("r-prof/proffer")The proffer package requires the RProtoBuf package, which may
require installation of additional system dependencies on Linux. See its
installation
instructions.
proffer requires the copy of pprof that comes pre-packaged with the
Go language. You can install Go at https://go.dev/doc/install.1
You can set the PROFFER_GO_BIN environment variable to a custom
location for the Go binary. See
usethis::edit_r_environ()
for directions on how to make this configuration permanent.
Run pprof_sitrep() again to verify that everything is installed and
configured correctly.
library(proffer)
pprof_sitrep()
#> • Call test_pprof() to test installation.
#>
#> ── Requirements ────────────────────────────────────────────────────────────────
#> ✔ Go binary '/usr/local/go/bin/go'
#>
#> ── Custom ──────────────────────────────────────────────────────────────────────
#> ✔ `PROFFER_GO_BIN` '/usr/local/go/bin/go'If all dependencies are accounted for, proffer should work. Test it
out with test_pprof(). On a local machine, it should launch a browser
window showing an instance of pprof.
library(proffer)
process <- test_pprof()When you are done testing, you can clean up the process to conserve resources.
process$kill()Recent versions of Go implement telemetry by default. Functions in
proffer such as pprof() turn off telemetry in order to comply with
CRAN policies. Read https://go.dev/doc/telemetry to learn how to
restore telemetry settings after using proffer.
We encourage participation through
issues and pull
requests. proffer has a
Contributor Code of
Conduct.
By contributing to this project, you agree to abide by its terms.
Profilers identify bottlenecks, but the do not offer solutions. It helps to learn about fast code in general so you can think of efficient alternatives to try.
- http://adv-r.had.co.nz/Performance.html
- https://www.r-bloggers.com/2016/01/strategies-to-speedup-r-code/
- https://www.r-bloggers.com/2013/04/faster-higher-stonger-a-guide-to-speeding-up-r-code-for-busy-people/
- https://cran.r-project.org/package=data.table/vignettes/datatable-intro.html
The profvis package is easier to
install than proffer and easy to invoke.
library(profvis)
profvis({
n <- 1e5
x <- data.frame(x = rnorm(n), y = rnorm(n))
for (i in seq_len(n)) {
x[i, ] <- x[i, ] + 1
}
x
})However, profvis-generated flame graphs can be difficult to
read and slow to respond
to mouse clicks.
proffer uses pprof to create
friendlier, faster visualizations.
Footnotes
-
One of the graph visualizations requires Graphviz, which you https://www.graphviz.org/download, but this visualization is arguably not as useful as the flame graph. ↩

